

 Skip to navigation
Skip to navigation
Site Primary Navigation:
- About SDSC
- Services
- Support
- Research & Development
- Education & Training
- News & Events
Search The Site:

Published June 28, 2018

Supercomputers helped study the binding of a plastic-degrading enzyme, PETase, which could lead to developing industrial-scale plastic recycling for throw-away bottles and carpet. Electrostatic potential distribution shown of PETase structure. Image courtesy of Gregg Beckham.
By Jorge Salazar
A dump truck's worth of plastic empties into the ocean every minute. Worldwide, humankind produces over 300 million tons of plastic each year, much of which is predicted to last centuries to millennia and pollutes both aquatic and terrestrial environments.
PET plastic, short for polyethylene terephthalate, is the fourth most-produced plastic, used to make things such as beverage bottles and carpets, most of which are not being recycled. Some scientists are hoping to change that, using supercomputers to engineer an enzyme that breaks down PET. They say it's a step on a long road toward recycling PET and other plastics into commercially valuable materials at industrial scale.
“We're ideally going from a place where plastics are hard to recycle to a place where we use nature and millions of years of evolution to direct things in a way that make plastic easy to recycle,” said Lee Woodcock, an associate professor of chemistry at the University of South Florida. Woodcock co-authored a study on the structure of an enzyme to degrade PET that was published in the Proceedings of the National Academy of Sciences (PNAS) in March 2018.
The study builds on a discovery in 2016 by Yoshida et al. of a bacterium, Ideonella sakaiensis 201-F6, which feeds on PET plastic as its source of carbon and energy. The PNAS study authors focused on the bacteria’s plastic-degrading enzyme, called PETase. Team members at the University of Portsmouth, led by Professor John McGeehan, used X-ray crystallography at the Diamond Light Source in the United Kingdom to solve the high resolution crystal structure of PETase.

Electron microscope images showing interaction of PETase enzyme with PET plastic. Image courtesy of Gregg Beckham.
“We then used computer simulations to understand how a polymeric ligand like PET would be able to bind to the enzyme,” said study co-author Gregg Beckham, a Senior Research Fellow and group leader at the US National Renewable Energy Laboratory (NREL). “We also conducted experimental work to show that indeed, the PETase can break down water or soda bottles, industrially relevant PET films, and another plastic, polyethylene furanoate.”
After doing this work on the structure and function of the PETase enzyme, the authors next tried to understand its evolution and look to a similar enzyme, a family of cutinases, which degrade the waxy polymer cutin found on the surface of plants.
“We developed the hypothesis that if we make the PETase enzyme more like a cutinase, then we should make the enzyme worse. When we did this work, in fact we ended up making the enzyme slightly better by doing that,” Woodcock said.
“It was incredibly surprising to us,” Beckham explained. “When we made it more cutinase-like, the enzyme was modestly improved. That's actually one of the key aspects of where computation came in, because it allowed us to essentially predict or suggest that aromatic-aromatic interactions in the enzyme with the aromatic polyester PET could potentially be responsible for its improved activity. But it was quite a surprise to us.”
Supercomputers allowed them to tackle tough science questions on PETase, such as the details of how it interacts on a molecular scale bound to a substrate, something beyond the scope of what could be determined by knowing its crystal structure.
The researchers took advantage for this study of computational resources of XSEDE, the Extreme Science and Engineering Discovery Environment, funded by the National Science Foundation.
The researchers simulated the long timescales of the enzyme using the Chemistry at Harvard Macromolecular Mechanics (CHARMM) force field and program itself, as well as Nanoscale Molecular Dynamics (NAMD) software. They used Comet at the San Diego Supercomputer Center (SDSC) at the University of California San Diego, as well as the Stampede1 and Stampede2 systems at the Texas Advanced Computing Center (TACC).
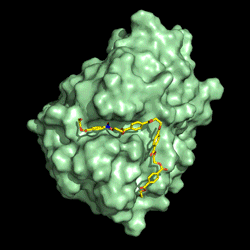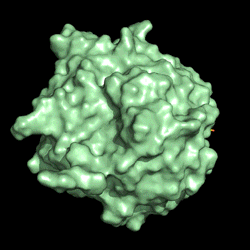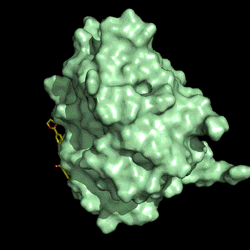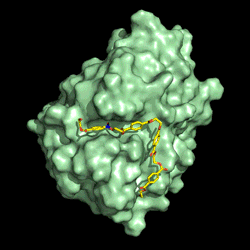
Electrostatic potential distribution of PETase structure. Image courtesy of Gregg Beckham.
“One nice thing about Comet, is that you have, for jobs that you need to get through in a high-throughput fashion, a shared queue which allows you to submit much smaller jobs but do it in a very high-throughput fashion, as they can share cores on the nodes,” said Woodcock. “This was particularly helpful.”
“We're producing a lot of great science across the spectrum of what our groups are collectively doing together using Stampede2 right now,” added Beckham. “Certainly, for the research on the plastics-degrading enzyme, we're using it for manuscripts and studies going forward on this same topic.”
Beckham said that their work has just begun on enzymes that clean up plastic pollution. “We're just starting to understand how this enzyme has evolved,” he said, adding that he wants to use computation to take advantage of large databases of genomics and metagenomics on enzymes to find the needles in the haystack that can degrade plastics.
The researchers are also interested in finding out of they can accelerate the degradation of PET at much higher temperatures, which could be industrially relevant in terms of using an enzyme to degrade PET and then convert that into the higher value materials, which could incentivize higher rates of reclamation, especially in the developing world where lots of plastic waste goes into the ocean.
“Understanding how we can better design processes to recycle plastics and reclaim them is a dire global problem and it's something (for which) the scientific and engineering community has to come up with solutions,” said Beckham.
Additional authors of the study, called ‘Characterization and engineering of a plastic-degrading aromatic polyesterase,’ include Harry P. Austin, Mark D. Allen, Alan W. Thorne, and John E. McGeehan (University of Portsmouth); Bryon S. Donohoe, Rodrigo L. Silveira, Michael F. Crowley, Antonella Amore, Nicholas A. Rorrer, Graham Dominic, William E. Michener, and Christopher W. Johnson, (National Renewable Energy Laboratory); Fiona L. Kearns and Benjamin C. Pollard (University of South Florida); Munir S. Skaf (University of Campinas); Ramona Duman, Kamel El Omari, Vitaliy Mykhaylyk, and Armin Wagner (Diamond Light Source, Harwell Science and Innovation Campus). The National Renewable Energy Laboratory Directed Research and Development Program funded the study, with computer time provided by the Extreme Science and Engineering Discovery Environment (XSEDE) under allocation MCB-090159.
About SDSC
As an Organized Research Unit of UC San Diego, SDSC is considered a leader in data-intensive computing and cyberinfrastructure, providing resources, services, and expertise to the national research community, including industry and academia. Cyberinfrastructure refers to an accessible, integrated network of computer-based resources and expertise, focused on accelerating scientific inquiry and discovery. SDSC supports hundreds of multidisciplinary programs spanning a wide variety of domains, from earth sciences and biology to astrophysics, bioinformatics, and health IT. SDSC’s petascale Comet supercomputer is a key resource within the National Science Foundation’s XSEDE (Extreme Science and Engineering Discovery Environment) program.
Share