

 Skip to navigation
Skip to navigation
Site Primary Navigation:
- About SDSC
- Services
- Support
- Research & Development
- Education & Training
- News & Events
Search The Site:

| BIOLOGICAL MODELING | Contents | Next
|
|
Determining Why Nature Needs Cholesterol |
|
| Featured Alexander Smondyrev Max Berkowitz University of North Carolina |
|
|
CHOLESTEROL IN THE MEMBRANEMOLECULAR DYNAMICS SIMULATIONSREFINING AMBERREFERENCES Smondyrev, A.M., Smondyrev, A,M., Smondyrev, A.M., and M.L. Berkowitz. 1999. Molecular dynamics simulation of dipalmitoylphosphatidylcholine membrane with cholesterol sulfate. Biophysical Journal (submitted). |
|
|
|
 |
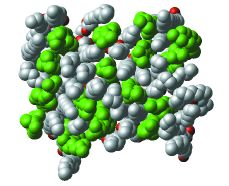 |
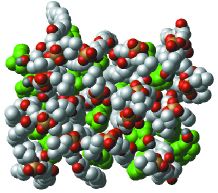
Figure 1. The headgroups of DPPC-cholesterol bilayerIn a membrane, the headgroups of DPPC molecules cover the molecules of cholesterol. Cholesterol carbon atoms are shown in green. Among DPPC atoms, phosphate is shown in orange and phosphate group oxygens are in red. |
Figure 3. Lipid bilayers with low cholesterol contentIn a side view of a lipid bilayer with low cholesterol content, the tails of DPPC molecules are disordered. Cholesterol molecules are shown in green, water as blue wires, and DPPC atoms as balls and black wires for the hydrocarbon chains. At high sterol content, the DPPC tails are more rigid. |
|
|
|
CHOLESTEROL IN THE MEMBRANEThe initial placement of cholesterol molecules in lipid membranes presented a challenge to the researchers. Because they were not able to observe diffusion in time periods long enough for two molecules in a lipid membrane to exchange their positions, they needed to start with a guess about the structure of the membrane with cholesterol. Previous studies suggested two possible packing structures for equal mixtures of DMPC and cholesterol. One favors "like-like" interactions such as lipid-lipid and cholesterol-cholesterol, and another "like-unlike," such as lipid-cholesterol. Both predicted patterns have similar energies. The researchers performed two simulations with equal mixtures of DPPC and cholesterol bilayers, assuming both like-like (Figures 1 and 2) and like-unlike packing. Although certain differences in membrane structure were observed, it was clear that longer simulations were needed to test this hypothesis directly. Of the various simulations performed by Smondyrev and Berkowitz, probably the most compelling was the molecular dynamics simulation of a DPPC membrane with another sterol, cholesterol sulfate. A paper describing the details of this simulation will be published later this year in Biophysical Journal. Although the structures of cholesterol and cholesterol sulfate are similar, they have different functions in biological systems.While cholesterol regulates membrane fluidity, membrane permeability, and lateral mobility of proteins, cholesterol sulfate acts as a membrane stabilizer. This difference may be due in part to different polar headgroups of the two molecules. The headgroup of cholesterol is a small, polar hydroxyl group, while cholesterol sulfate has a large, charged, and hydrated polar headgroup. Cholesterol sulfate is found in spermatozoon plasma membrane, stratum corneum (the outermost layer of the skin), and the membranes of red blood cells. For example, past research shows that cholesterol sulfate helps protect the membranes of red blood cells from destruction by lysins--antibodies that disintegrate red blood cells or bacteria cells. It is also believed that cholesterol sulfate has a noticeable effect on fertilization efficiency. While cholesterol sulfate accounts for only 2% of the total sterol in the human sperm, its concentration in the membranes overlying the acrosome--the head area that contains enzymes to digest the egg cell coating--is as much as 20%. It is likely that cholesterol sulfate contributes to membrane stability in this region. Smondyrev and Berkowitz studied the changes occurring in a DPPC-cholesterol membrane at a high sterol content. In their previous studies, they studied low amounts versus high amounts of cholesterol, with results matching experimental data. Now, they replaced cholesterol with cholesterol sulfate. Again, the results agreed with data from experiments. |
Top | Contents | Next |
|
|
|
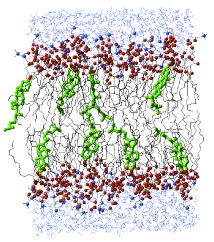 |
Figure 3 - Lipid Bilayers with Low Cholesterol Content
In a side view of a pilip bilayer with low cholesterol content, the tails of the DPPC molecules are disordered. Cholesterol molecules are shown in green, water as blue wires, and DPPC atoms as balls and black wires for the hydrocarbon chains. At high sterol content, the DPPC tails are more rigid. |
|
|
|
MOLECULAR DYNAMICS SIMULATIONSMuch of the study on sterol-containing membranes was done on the Cray T3E at SDSC and the Cray T3E at the Texas Advanced Computing Center at the University of Texas at Austin, an NPACI resource partner. Calculations were also performed on a Cray T3E at the North Carolina Supercomputer Center. Smondyrev and Berkowitz applied molecular dynamics simulation techniques to study the structural and dynamic properties of the membranes. This technique was a well-established tool in the study of chemical dynamics, electronic structure, structure of condensed matter, and structure and dynamics of such biomolecules as protein and DNA. In studying membranes, this technique allowed them to explore some of the characteristics, such as permeability or surface potentials, that are hard to obtain using other methods. They used the parallel molecular dynamics simulation package DL_POLY, which was developed at Daresbury Laboratory by W. Smith and T.R. Forester. It performs molecular dynamics in many different ensembles, using a wide variety of algorithms for time-stepping, electrostatic models, periodic images, and rigid molecular entities. The DL_POLY force field includes a range of features. The parameterization can be obtained, for example, from AMBER or GROMOS force fields. It is also easy to adapt DL_POLY to user-specific force fields. To reduce the computational intensity of the problem, they used the united atom model. In this model, CH2 and CH3 groups are treated as a single, "united" atom. This model is especially useful in problems concerned with the modeling of very large systems, such as proteins in lipid membranes. Force fields for united atom models are included in such packages as AMBER and GROMOS, but these were originally designed to describe proteins and nucleic acids. AMBER had to be modified to simulate sterols in membranes. |
Top | Contents | Next |
REFINING AMBERSmondyrev and Berkowitz refined the united atom AMBER force field for the simulation of phospholipid membranes. This was done by obtaining an accurate set of torsional parameters corresponding to a set of atomic partial charges calculated for DPPC and DMPC molecules. Bond and angle parameters were adopted from the AMBER force field, while torsional parameters were fitted to reproduce geometry and energy profiles obtained from ab initio calculations of model compounds. The Ryckaert-Bellemans potential was employed to describe the torsion potentials for the united atoms in lipid chains. To test the force field, Smondyrev and Berkowitz performed a constant pressure simulation on a DPPC bilayer in water (Figure 3). The average area per headgroup, peak-to-peak distance in the electron density, and order parameter profile for the hydrocarbon chains agreed with the experimental values and with the results of a simulation using a model with explicit hydrogens. "Typical cell membranes contain mixtures of phospholipid molecules with proteins and sterols," Smondyrev said. "The long-term goal of our work is to develop more realistic models of biological membranes. Then we can study mechanisms by which cholesterol influences the properties of proteins and channels in membranes. Eventually, we may shed some light not only on how cholesterol works in the membranes of the human body, but why such a potentially hazardous compound is there in the first place." |
Top | Contents | Next |