SCIENCE OF STRUCTURE DETERMINATION
A YEAR OF DISCOVERY
THE ROLE OF BIOINFORMATICS
 cientists have sequenced the genomes of more than 800 organisms, and as the list grows researchers in California and elsewhere are racing to understand what the tens of thousands of proteins encoded by the DNA blueprints actually do. The crucial first phase of that formidable task is structural genomics, the process of determining the 3-D structure of all the proteins to gain insights into their functions and interrelationships. Structural genomics will enable researchers to understand how flawed proteins cause diseases and how to design drugs to treat those diseases. cientists have sequenced the genomes of more than 800 organisms, and as the list grows researchers in California and elsewhere are racing to understand what the tens of thousands of proteins encoded by the DNA blueprints actually do. The crucial first phase of that formidable task is structural genomics, the process of determining the 3-D structure of all the proteins to gain insights into their functions and interrelationships. Structural genomics will enable researchers to understand how flawed proteins cause diseases and how to design drugs to treat those diseases.
 |
Figure 1. Handling Crystals Robotically
A device at the lower end of this robotic arm withdraws a crystal from a canister before placing it into a beamline at the Stanford Synchrotron Radiation Laboratory. The diffraction of X-rays passing through the crystal reveals underlying molecular structure. |
In recognition of the enormous potential to benefit humanity, the National Institutes of Health (NIH) has funded nine multi-institution consortia seeking to determine protein structures with assembly-line approaches (Figure 1). One of these "high-throughput" projects, the Joint Center for Structural Genomics (JCSG), involves SDSC and UCSD, The Scripps Research Institute, the Genomics Institute of the Novartis Research Foundation (GNF), Stanford University and the Stanford Synchrotron Radiation Laboratory, and the Salk Institute. The NIH’s National Institute of General Medical Sciences has awarded five-year grants to the nine projects as part of its Protein Structure Initiative. "Our ambitious initiative in the United States is part of a worldwide effort," said John Norvell, program director for the Protein Structure Initiative.
Major structural genomics consortia have formed in Japan, Germany, the United Kingdom, and Canada, with other efforts under way in Sweden, France, Italy, Brazil, and China. The International Structural Genomics Organization was founded in 2001 to coordinate the goals of all the projects. The U.S. effort is focused on finding the structures for representatives of all the families of proteins and protein "folds."
SCIENCE OF STRUCTURE DETERMINATION
 |
|
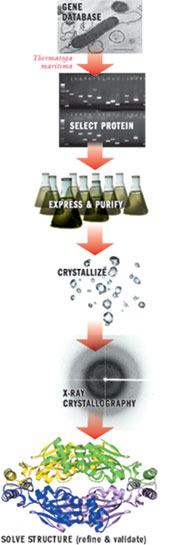
Figure 2. The Structural Genomics Pipeline
The Joint Center for Structural Genomics cloned a specific gene (TM0449) of the bacterium Thermotoga maritima, purified and crystallized the protein, and exposed crystals to an X-ray beamline as part of a high-throughput process of determining the 3-D structures of proteins.
|
Proteins are strings of amino acids. The order of the amino acids along the string can be determined by several techniques, but the 3-D structure of any protein is hard to predict because amino acids distant from one another often come in close contact. An understanding of proteins in living cells is possible only when their 3-D structures have been determined experimentally, revealing folds and other features. "The goal is to fully yet efficiently characterize the range of protein folds," said Norvell.
A previously unknown protein fold, which also is a possible new drug target, was among the first findings in JCSG’s highly promising past year. "We’ve hit the ground running," said project director Ian Wilson of Scripps. "There are more surprises in store."
The objectives of JCSG are being carried out in a systematic, step-by-step fashion (Figure 2). "Efficient coverage of families and fold space requires strategic selection of target proteins for solution," said Susan S. Taylor, a professor of chemistry and biochemistry at UCSD, chairperson of JCSG’s target selection effort, and a member of the National Academy of Sciences. With global coordination, she said, the community could minimize the number of structure solutions required to serve as templates for solving others.
"Success in these projects will change the science of structure determination," said Wilson. The advantages will include a tenfold reduction in the cost per protein solution, a vast increase in data available to biomedical scientists, and an ability to answer more complex questions. "We will be able to ask not only ‘What does this protein do?’ as we can today," Wilson said, "but also ‘What do all these proteins do collectively?’ or ‘Why do some metabolic pathways of protein activity occur in some organisms and not others?’ and ‘How might we specifically interfere with them in microbial pathogens?’–questions at a more holistic level."
A YEAR OF DISCOVERY
"This year we have identified and, in many cases, opened up several bottlenecks in the process of high-throughput structure determination, but not all of them," said Raymond Stevens, leader of the JCSG Crystallomics Core at Scripps. Stevens and Peter Schultz, also at Scripps, are speeding up structural genomics with robotics designed for rapid expression, purification, and crystallization of proteins.
Some of the first structures solved by JCSG are produced by the bacterium Thermotoga maritima, one of the planet’s oldest life-forms, which was discovered in geothermally heated marine sediments. The bacterium’s 1,877 genes code for relatively small proteins that remain active at 80° C and crystallize relatively easily in the laboratory. "Indeed, we chose this system because it provides an opportunity to do structural genomics on an entire organism, and it enables an ideal end-to-end test of our system," said Wilson.
The GNF team, led by Scott Lesley, successfully amplified 1,791 of the 1,877 T. maritima genes in the high-throughput system, and 1,369 were cloned and expressed in a host bacterium. The proteins were purified, then crystallized, a necessary step for structure determination by X-ray crystallography.
Historically, crystallization of a protein has been a trial-and-error enterprise that occasionally took years. Automation and miniaturization allows crystals to form faster from smaller quantities of protein. The second-generation robotic crystallization system developed by Stevens and engineers at GNF and Syrrx, Inc., can produce 60,000 crystallization trials a day, across a range of temperatures and other conditions. The system uses "nanodrops" (50 nanoliters) of protein solution. A robot scans all trials for crystal formation. By January 2002, the T. maritima trials had produced more than 300 high-quality crystals.
The crystals are sent to Peter Kuhn, who leads the Structure Determination Core at the Stanford Synchrotron Radiation Laboratory. Kuhn and his colleagues at the laboratory have designed and built an automated sample-changing system (Figure 1) to select, characterize, and transfer premounted crystals grown in nanodrops onto the synchrotron’s X-ray beamline for analysis. Data sets can be taken for up to 285 crystals without further human intervention.
In a beamline, X-rays pass through each crystal, a repetitive array of protein molecules, producing a diffraction pattern of numerous "reflections," sets of X-ray spots. The position and intensity of the spots depend on the type and 3-D arrangement of atoms in the crystal. Many reflections are collected for each crystal and examined computationally to produce a structural solution: a map of the distribution of each atom in space. Each solution is further validated, and researchers add the final 3-D coordinates to the Protein Data Bank, the international repository of protein structures.
One of the JCSG’s solved protein structures is TM0449, an enzyme needed by T. maritima for replication and repair of its DNA. The enzyme has a completely novel protein fold, which has been added to the Protein Data Bank and international databases of protein topologies. "The significance is that, in pursuit of presumably simple proteins that have survived as long as life has been on Earth, we find that we can still discover something new," said John Wooley, a JCSG co-principal investigator and associate vice chancellor for research at UCSD. The new protein fold was also found in a number of pathogens (including the deadly anthrax bacterium). The molecule may become a target for new antimicrobial drugs.
THE ROLE OF BIOINFORMATICS
In addition to miniaturization and process automation, bioinformatic technologies are making high-throughput genomics faster and more informative. "It isn’t just quantity we’re after," said Adam Godzik, leader of JCSG’s Bioinformatics Core, located at SDSC. "We make the whole process faster because our database records successes and failures at each stage, information that enables us to raise the success rate. Our processes and results are accessible on our website." As required by the International Structural Genomics Organization, all U.S. structural genomics projects have target lists accessible on the Web. Each project is committed to rapid deposition (within 4—6 weeks after completing refinement) of new structures in the Protein Data Bank.
"We have been rolling out variations on existing tools and developing new ones to aid the high-throughput model," said Godzik. On the website, homology and structure analysis tools are augmented by other tools, including a Data Acquisition Prioritization System. The location of the Bioinformatics Core at SDSC also allows JCSG to take advantage of SDSC’s resources and expertise in manipulating large data sets.
"We have begun on a very promising note," said Wilson. "Our efforts are now complementary to those pursuing structures one by one: we all find that the X-ray structure solutions often suggest functional roles for proteins. High-throughput structural biology will also facilitate comprehensive studies of complete metabolic pathways. It will lift all our questions onto a philosophically higher plane. We will be able to find out how complex assemblies of protein components provide a blueprint for the operation and function of a cell. Protein structure determination, per se, will no longer be the main obstacle in elucidating the molecular basis of biology." –MM
|
Principal Investigator
Ian Wilson
The Scripps Research Institute
Co-Principal Investigators
John Wooley
UCSD
Keith Hodgson
Stanford University, the Stanford Synchrotron Radiation Laboratory
Participants
Adam Godzik
SDSC
Peter Kuhn
Stanford University, the Stanford Synchrotron Radiation Laboratory
Raymond Stevens
The Scripps Research Institute, the Genomics Institute of the Novartis Research Foundation
Susan S. Taylor
REFERENCES
R.C. Stevens, S. Yokoyama, and I.A. Wilson (2001): Global efforts in structural genomics, Science 294, 89-92.
R.C. Stevens and I.A. Wilson (2001): Industrializing structural biology, Science 293: 519-520.
|


 Skip to navigation
Skip to navigation
