

 Skip to navigation
Skip to navigation
Site Primary Navigation:
- About SDSC
- Services
- Support
- Research & Development
- Education & Training
- News & Events
Search The Site:

| ALPHA PROJECTS | Contents | Next | |
Scalable Visualization Toolkits |
|
| PROJECT LEADERS Arthur Olson, The Scripps Research Institute (TSRI) Bernard Pailthorpe, UCSD/SDSC Arthur Toga, UCLA Carl Wunsch, MIT |
PROJECT PARTICIPANTS Mick Fellows, John Marshall, MIT Nicole Bordes, Alex de Castro, Jon Genetti, Reagan Moore, John Moreland, David Nadeau, Arcot Rajasekar, SDSC Michel Sanner, TSRI Colin Holmes, Paul Thompson, UCLA Scott Baden, UC San Diego Detlef Stammer, UCSD/Scripps Institution of Oceanography Joel Saltz, University of Maryland/ Johns Hopkins University Chandrajit Bajaj, University of Texas Andrew Grimshaw, University of Virginia |
|
FROM GREY MATTER......TO THE DEEP BLUE SEAEXTENDING THE INTERACTION INFRASTRUCTUREACCESSING THE DATA |
|
|
|
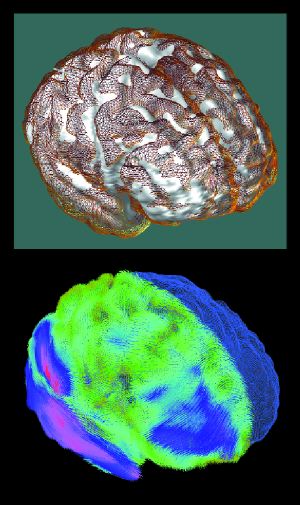
Figure 1. Mapping the Ravages of Alzheimer's Disease
LONI researchers are building population-based digital brain atlases to discover how brain structures are altered by disease. In 3-D maps of variability from an average cerebral cortex surface derived from 26 Alzheimer's disease patients, individual variations in brain structure are calculated based on the amount of deformation required to drive each subject's convolution pattern into correspondence with the group average. Surface matching figures are computed using fluid flow equations with more than 100 million parameters; this requires parallel processing and very high memory capacity. Image by Paul Thompson, Colin Holmes, and Arthur Toga, Laboratory of Neuro Imaging, UCLA. |
 |
|
|
|
FROM GREY MATTER..."In our brain mapping research, we're developing tools and data sets to measure the structure and function of both healthy and diseased brains," said Arthur Toga, director of UCLA's Laboratory of Neuro Imaging (LONI). "We're also integrating information among methods, populations, laboratories, and species, guided by problems that affect humans with neurological or psychiatric disorders." Toga's laboratory concentrates on four areas: the extraction and analysis of complex shape information from anatomic structures, the extraction and visualization of differences in brain shape using volume-based, nonlinear warping algorithms, modeling and correlating changes in gross features and landmarks with cellular-level characteristics, and developing visualization tools that accommodate not only spatial location and intensity but also time, in the form of animation. "Neuroimaging and brain mapping typically are conducted in 3-D," said UCLA assistant professor Paul Thompson, head of the LONI modeling team. "Our goal is to add the time dimension and develop a modeling environment that will extend the static measurement of the brain to a study of dynamic, evolving conditions as brains develop, age, or deteriorate as a result of pathological conditions such as Alzheimer's disease." The LONI researchers will provide multi-modal volumetric data of human brains and models, at hundreds of gigabytes per data set, as test cases for the Scalable Visualization Toolkits effort. |
Top| Contents | Next |
...TO THE DEEP BLUE SEACarl Wunsch at MIT and Detlef Stammer at UCSD's Scripps Institution of Oceanography (SIO) are modeling the general circulation of the ocean on a global, continuous basis. The MIT Ocean State Estimation project, a system for global data assimilation and analysis, analyzes World Ocean Circulation Experiment hydrographic and current data and TOPEX/ POSEIDON satellite observations in worldwide and regional models. "Our Global Circulation Model divides the surface regions of our planet into thousands of simulation cells," Wunsch said. "We keep track of the flow field, temperature, salinity, oxygen content, nutrients, carbon and nitrogen markers, and sea surface. The volume of data is overwhelming, and detecting important features in such large data sets is a challenging computational problem." In particular, their research models oceanic heat and fresh water fluxes to understand the relationships between ocean circulation and climate changes. The computational fluid dynamics simulations can produce 100-gigabyte data sets, which will be test cases for the Scalable Visualization Toolkits effort. "Our simulation results are important for understanding and predicting the extent of global climate change," Stammer said. "Our colleagues Mick Fellows and John Marshall also use the model results to calculate oceanic biological fluxes and how they relate to ocean color, biological tracers, and the carbon cycle, all of which affect ecosystems as well as climate." |
Top| Contents | Next |
EXTENDING THE INTERACTION INFRASTRUCTUREThe alpha project will leverage renderers, user interfaces, and data-access utilities from various Interaction Environments projects into a coherent whole. "NPACI is fortunate to have many of the pieces of the puzzle already in place," Pailthorpe said. "Nevertheless, organizing, extending, and integrating these tools won't be a trivial task." SDSC's David Nadeau is developing a space-time expression tree, a general infrastructure to organize data flow and computation for large visualization tasks that can be viewed as the core of the toolkit. He already is working with elements of the system that perform data manipulation and translation, support composition of "3-D+time" data elements, and provide high-performance interfaces to the tools within parallel rendering applications. Two ongoing NPACI projects complement the needs of the scientific applications for rendering volume and surface data. SDSC's Massively Parallel Interactive Rendering Environment (MPIRE) is a volume renderer that creates images from 3-D voxel-based data sets on supercomputers. Developed by SDSC's Jon Genetti and Greg Johnson, MPIRE has been applied to problems ranging from medical imaging to fluid dynamics to rendering astronomical objects (see p. 18). It can be used interactively from any computer on the Web via Java applets. MPIRE recently has been extended to include 3-D perspective and soon will be enhanced to deal with additional formats and to render terabyte-scale data sets too large to fit in memory. At the Center for Computational Visualization of the Texas Institute for Computational and Applied Mathematics, Chandrajit Bajaj leads the VisualEyes project, which is building analysis and visualization tools for large simulations. VisualEyes analysis tools analyze sets of sequential 2-D images, and from them extract 3-D geometry data. Fast isocontouring tools analyze physical features--flow, temperature, or stress, for example--in simulation results and plot isocontours on a 3-D mesh, so points on the mesh with the same physical values can be visualized with the same colors and transparency values. Scalar and vector topology analysis methods identify points, lines, and 2-D features that mark transitions in 3-D data sets. Hypervolume rendering methods reduce 3-D or higher-dimensional results to 2-D representations. Coupling the rendering operation to a graphical user interface allows users to change interactively the projection parameters to explore multi-dimensional data and identify regions of interest. "Voxel-based MPIRE and surface-oriented VisualEyes are beautifully complementary," Pailthorpe said. In the Active Data Repository (ADR) project, researchers are working with Genetti and Pailthorpe to develop efficient raycasting algorithms to render very large datasets. But the tools that support 3-D rendering also perform many other tasks that involve processing and manipulation of very large datasets. |
Top| Contents | Next |
ACCESSING THE DATAJoel Saltz, the leader of NPACI's Programming Tools and Environments thrust area, heads the ADR project at the University of Maryland. ADR optimizes storage, retrieval, and processing of very large multi-dimensional data sets and supports common database operations, data retrieval, memory management, scheduling of processing , and user interaction. The system provides a uniform interface to the data and processing; to customize it for various applications, an application developer need only provide a data description and the processing methods. The ADR already is being applied to projects in geography, medicine, and environmental science. In a collaboration with Mary Wheeler's group at the University of Texas, researchers linked separate water flow and chemical transport simulation codes to accurately model pollutant transport in bays and estuaries. The ADR handled multi-gigabyte data files and reconciled incompatible data formats. Under the Scalable Visualization Toolkits effort, the ADR server will be ported from an IBM SP at the University of Maryland to the SP at SDSC and extended to use tape archives, such as the High Performance Storage System (HPSS). Large data sets will be accessed via an applications program interface to the Storage Resource Broker (SRB), developed at SDSC under the direction of Regan Moore. The compute-intensive rendering and analysis components of the toolkit will use distributed parallel execution capabilities developed under the NPACI Metasystems thrust. Andrew Grimshaw's group at the University of Virginia will guide the use of Legion to distribute applications across NPACI resources. A separately funded effort by UC San Diego's Scott Baden will enhance and adapt the KeLP parallel programming system. Arthur Olson and Michel Sanner at The Scripps Research Institute will direct client-side integration of the visualization toolkits. Sanner's Déjà Vu interface gives users viewing and manipulation capabilities and will be used as a prototype. Based on Python, an interpreted, interactive, object-oriented programming language comparable to Perl and Java, Déjà Vu is portable to many computers and operating systems. SDSC's John Moreland will integrate the visualization toolkit with the ICE collaborative environment, of which MICE (Molecular ICE) is the best-known implementation. "The Scalable Visualization Toolkits effort is a high-payoff project," Pailthorpe said. "A 'plug-and-play' visualization environment, with modular tools that scale from desktop computers to the PACI Grid, will be useful to all disciplines within NPACI and will assist researchers throughout the nation." --MG * |
Top| Contents | Next |